Milestone Achievement
We have reached a new milestone by implementing tests for key MultiXscale applications in the EESSI test suite.
The EESSI test suite is a suite of portable application tests, implemented in the ReFrame HPC testing framework. While primarily designed to test software in the EESSI software environment, it can be run on any (module-based) software environment, such as those typically provided by HPC sites on their systems.
The test suite scans available modules, and generate tests for those modules for which a test case has been implemented. A range of important applications in EESSI was already covered in the test suite. The current milestone reflects that EESSI test suite, as of version 0.9.0 (https://github.com/EESSI/test-suite/releases), covers all the key applications developed in WPs 2, 3 and 4 of MultiXscale with a test case, as they are described below.
The lbmpy [1] software package is a python-based domain specific language which automatically generates performance portable implementations of key Lattice Boltzmann (LB) ingredients such as velocity space discretizations, equilibrium and boundary representations, collision models, etc. In this framework, a typical MPIX parallelized LB simulation workflow couples highly optimized shared memory parallelized CPU/GPU kernels generated by lbmpy with the MPI parallel waLBerla [2] software.
From an LB domain scientists’ perspective, lbmpy offers a convenient python interface wherein new developments can be quickly prototyped, and preliminary scientific validations can be conducted via OpenMP/CUDA parallelized tests. Nevertheless, the utility of lbmpy is particularly limited to moment-space LB methods which, as compared to traditional population-space methods, offer better stability characteristics albeit at the cost of computational efficiency. However, extensive research has been recently conducted to improve the stability of population-space LB methods. Indeed, such research has culminated in the development of several advanced population-space models.
Consequently, lbmpy has been extended to also allow population-space descriptions and is available in the multixscale-lbmpy [3] repository. At present the traditional Bhatnagar-Gross-Krook collision model is validated for a test case [4] that simulates the Kelvin-Helmholtz instability where an initial hyperbolic tangent velocity profile imposed in a fully periodic 2D square box is slightly perturbed to initiate rolling of the shear layers. The test conducts a validation run and computes the normalized kinetic energy as a validation metric. Further, the performance of the employed stream-collide algorithm is evaluated and reported in Mega lattice updates per seconds (MLUPS). The test can be executed in 2 modes, namely, serial/openmp parallelized.
- Serial runs:
python mixing_layer_2D.py - OpenMP parallel runs:
OMP_NUM_THREADS=4 python mixing_layer_2D.py –openmp
Efficient load balancing is critical for large-scale particle simulations on modern HPC systems as the computational workload and data distribution can change significantly over time due to particle motion, interactions and adaptive resolution. Without dynamic load balancing, these effects result in imbalances in the workload across processes, reduced `parallel efficiency and poor scalability at extreme core counts.
The A Load Balancing Library (ALL) addresses these challenges by providing domain decomposition load-balancing strategies tailored to particle-based applications. By optimizing the domain decomposition through a range of available methods, ALL helps to maintain an even workload distribution, minimise communication overhead and improve time-to-solution on heterogeneous and massively parallel architectures.
Within this milestone, we report on the integration and coupling of ALL with the LAMMPS particle simulation code. This coupling demonstrates how ALL can be applied to a production-grade molecular dynamics application, enabling improved scalability and performance on EuroHPC systems and highlighting the benefits of a reusable load-balancing library within a Centre of Excellence software ecosystem.
In addition, a coupled build integrating LAMMPS, ALL, and OBMD. OBMD simulations are characterized by dynamically changing particle populations and strongly non-uniform spatial workloads due to particle insertion, removal, and fluxes across open boundaries. In this context, efficient load balancing is particularly critical to sustain scalability and numerical efficiency, as imbalances can rapidly arise during the simulation. The integration of ALL with LAMMPS and OBMD demonstrates how adaptive load-balancing capabilities enable robust and efficient execution of advanced open-boundary particle simulations.
ALL souce code can be found at https://gitlab.jsc.fz-juelich.de/SLMS/loadbalancing, while examples using the coupling to LAMMPS are publicly available at: https://github.com/yannic-kitten/lammps/tree/ALL-integration/examples/PACKAGES/allpkg.
ESPResSo is a versatile particle-based simulation package for molecular dynamics, fluid dynamics and Monte Carlo reaction schemes [1,2]. It provides numerical solvers for electrostatics, magnetostatics, hydrodynamics, electrokinetics, and diffusion-advection-reaction equations. It is designed as an MPI-parallel and GPU-accelerated simulation core written in C++ and CUDA, with a scripting interface in Python which integrates well with science and visualization packages in the Python ecosystem, such as NumPy and PyOpenGL. Its modularity and extensibility made it a popular tool in soft matter physics, where it has been used to simulate ionic liquids, polyelectrolytes, liquid crystals, colloids, ferrofluids and biological systems such as DNA and lipid membranes. Some research applications lead to the development of specialized codes that use ESPResSo as a library, such as molecular builders (pyMBE [3], pyOIF [4], pressomancy [5]), reinforcement learning frameworks (SwarmRL [6]) and systematic coarse-graining frameworks (VOTCA [7,8]).
Many soft matter systems are characterized by physical properties that resolve different time- or length-scales. To fully capture these effects in a simulation, a multiscale approach is needed. Often, long-range interactions can be resolved very efficiently with minimal loss of accuracy using grid-based solvers, such as the particle–particle–particle mesh (P3M) algorithm for electrostatics and magnetostatics. Similarly, solute–solvent interactions can be approximated using the lattice-Boltzmann (LB) method for hydrodynamics, where the dense solvent is discretized on a grid and exchanges momentum with solid particles that represent the solute. These techniques not only bring the computational costs down, they also consume several orders of magnitude less computer memory compared to an atomistically-resolved system and require less bandwidth during communication between HPC nodes.
Choice of simulation scenarios
We opted for three main simulation scenarii that are described in more detail in Deliverable 2.1 [9]. They have been written in a backward-compatible way, such that ESPResSo releases 4.2 and 5.0 can execute them, despite the API changes between both versions.
The Lennard-Jones (LJ) scenario consists of soft spheres interacting with a short-range potential [10]. This simple setup underpins most particle-based simulations, where a weakly attractive, strongly repulsive pairwise potential is required to prevent particles from overlapping with one another. In addition, solvated systems with atomistic resolution typically have a large excess of solvent atoms compared to solute atoms; thus, LJ interactions tend to account for a large portion of the simulation time. The test simulates a Lennard-Jones fluid in the NVT ensemble and compares its pressure and total energy against expected values for the given volume, temperature, and particle number.
The ionic crystal scenario (P3M) builds upon the LJ scenario and adds an electrical charge to the particles [11]. Since the Coulomb interaction decays with the inverse of the distance, it is necessary to include periodic image contributions to the particle forces. This is achieved using long-range numerical solvers, which leverage the fast Fourier transform (FFT) algorithm to calculate forces in an efficient manner. This type of simulation finds many applications in soft matter research, such as ionic liquid-based batteries, supercapacitors, and polyelectrolytes (gelling agents, charged biomolecules). The test models an ionic crystal, whose electrostatic potential can be directly compared against the Madelung constant.
The hydrodynamics scenario is based on the LB method and models a dense, compressible fluid [12]. It was chosen due to the importance of hydrodynamic interactions in many physical systems at the mesoscale. While an atomistic description of solvent molecules is justifiable at the nanoscale, at the scale of micrometers a continuum-based description of the solvent is significantly less computationally demanding. It is also much more accurate than implicit solvent models, which do not adequately capture motion correlation in particles that are close to each other. With hydrodynamics and particle coupling, elements of fluid are resolved on a grid, allowing for momentum transfer between particles mediated via the fluid at the speed of sound. The test simulates a thermalized quiescent fluid with or without particles and verifies momentum conservation.
Running the testsuite
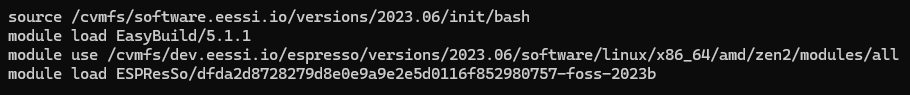
ESPResSo can be either compiled from sources using the instructions in the user manual [13] or loaded from the EESSI repository. Loading the latest version of ESPResSo from dev.eessi.io involves the following steps, assuming the workstation is equipped with an AMD microprocessor:
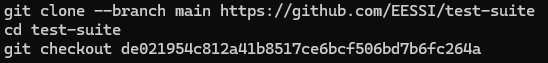
Tests are available on GitHub and can be downloaded with the following steps:
Tests can be executed as follows, assuming 2 MPI ranks and 4 OpenMP threads per MPI rank:
References:
[1] F. Weik, R. Weeber, K. Szuttor, K. Breitsprecher, J. de Graaf, M. Kuron, J. Landsgesell, H. Menke, D. Sean, and C. Holm, “ESPResSo 4.0 – an extensible software package for simulating soft matter systems,” Eur. Phys. J. Spec. Top., vol. 227, pp. 1789–1816, 2019
[2] R. Weeber, J.-N. Grad, D. Beyer, P. M. Blanco, P. Kreissl, A. Reinauer, I. Tischler, P. Košovan, and C. Holm, “ESPResSo, a versatile open-source software package for simulating soft matter systems,” in Comprehensive Computational Chemistry, 1st ed., M. Yáñez and R. J. Boyd, Eds. Oxford: Elsevier, 2024, pp. 578–601
[3] D. Beyer, P. B. Torres, S. P. Pineda, C. F. Narambuena, J.-N. Grad, P. Košovan, and P. M. Blanco, “pyMBE: the Python-based molecule builder for ESPResSo,” J. Chem. Phys., vol. 161, p. 022502, 2024
[4] I. Jančigová, K. Kovalčíková, R. Weeber, and I. Cimrák, “PyOIF: Computational tool for modelling of multi-cell flows in complex geometries,” PLoS Computational Biology, vol. 16, p. e1008249, 2020
[5] Mostarac, “Pressomancy,” GitHub. url:https://github.com/stekajack/pressomancy
[6] S. Tovey, C. Lohrmann, T. Merkt, D. Zimmer, K. Nikolaou, S. Koppenhöfer, A. Bushmakina, J. Scheunemann, and C. Holm, “SwarmRL: building the future of smart active systems,” Eur. Phys. J. E, vol. 48, 2025
[7] V. Rühle, C. Junghans, A. Lukyanov, K. Kremer, and D. Andrienko, “Versatile object-oriented toolkit for coarse-graining applications,” J. Chem. Theory Comput., vol. 5, pp. 3211–3223, 2009
[8] S. Y. Mashayak, M. N. Jochum, K. Koschke, N. R. Aluru, V. Rühle, and C. Junghans, “Relative entropy and optimization-driven coarse-graining methods in VOTCA,” PLOS ONE, vol. 10, pp. 1–20, 2015
[9] J.-N. Grad and R. Weeber, “Report on the current scalability of ESPResSo and the planned work to extend it,” EuroHPC Centre of Excellence MultiXscale, MultiXscale Deliverable 2.1, 2023
[10] LJ test pull request: https://github.com/EESSI/test-suite/pull/155. LJ test versioned link: https://github.com/EESSI/test-suite/blob/48d62e7/eessi/testsuite/tests/apps/espresso/src/lj/lj.py
[11] P3M test pull request: https://github.com/EESSI/test-suite/pull/144. P3M test versioned link: https://github.com/EESSI/test-suite/blob/48d62e7/eessi/testsuite/tests/apps/espresso/src/p3m/madelung.py
[12] LB test pull request: https://github.com/EESSI/test-suite/pull/300. LB test versioned link: https://github.com/EESSI/test-suite/blob/48d62e7/eessi/testsuite/tests/apps/espresso/src/lb/lb.py
[13] ESPResSo user manual, chapter on installation: https://espressomd.github.io/doc/installation.html
Ion adsorption and dynamics in porous carbons are crucial for many technologies, such as energy storage and desalination. Molecular simulations of electrolytes in contact with carbon-based materials, the most common approach to model such systems, is too computationally expensive to include the heterogeneity of experimental systems. In particular, materials usually have pore size and particle size distributions which cannot be represented in the relatively small systems simulated at the molecular level. LPC3D is a software designed for mesoscopic simulations of porous carbon particles and carbon-based supercapacitors which allow for the inclusion of such heterogeneity. The code calculates quantities of adsorbed ions, diffusion coefficients and NMR spectra of ions / molecules adsorbed in porous carbon matrices. In MultiXscale, we implemented a new version of LPC3D, written in Python using the PyStencils module which can generates optimized C++ code. This implementation is parallel, can be run on CPU and GPU, and allows one to simulate systems going from a single carbon particle to a supercapacitor with hundreds of micrometers in length.
For milestone, the system simulated is a porous carbon electrode, charged negatively, and containing BMIM-BF4 in acetonitrile as the electrolyte. The calculation of NMR spectra and diffusion coefficients are done for the anion. If the diffusion coefficients equals the desired value, the test is successful.
The code, a manual, and examples can be found in the following repository: https://github.com/multixscale/LPC3D. The version is 0.1.2.
Typically, molecular dynamics (MD) simulations are conducted in the canonical ensemble, which maintains a constant number of particles, volume, and temperature. However, many real systems are open and can exchange mass, momentum, and energy with their surroundings. Performing simulations of such systems requires the use of the grand-canonical ensemble, made possible by the OBMD method [1,2,3], which allows particle exchange through the opening of the system’s boundaries.
The OBMD method has been implemented within the Large-scale Atomic/Molecular Massively Parallel Simulator (LAMMPS) simulation package by adding a new OBMD package (commit efdde83). The latter includes obmd extension [4], which takes care of the deletion and insertion of particles, measures outflow of the linear momentum, and imposes external boundary conditions using external forces.
For milestone, we chose simulation of liquid water under equilibrium conditions as a test, which is described using the mesoscopic DPD water model [5]. The test checks the density of DPD water in the region of interest. If the density equals the desired value (within predetermined error), the test is successful.
Code can be found in the following repository: https://github.com/chocolatesoup/OBMD-LAMMPS-extension. The test presented can be found in the example directory of the LAMMPS code (commit e1de8e2, https://github.com/chocolatesoup/OBMD-LAMMPS-extension/tree/main/code/examples/OBMD/OBMD_DPD-EQUILIBRIUM).
References:
[1] R. Delgado-Buscalioni, J. Sablić, and M. Praprotnik, “Open boundary molecular dynamics,” Eur. Phys. J. Spec. Top., vol. 224, pp. 2331–2349, 2015
[2] L. Delle Site and M. Praprotnik, “Molecular systems with open boundaries: Theory and simulation,” Phys. Rep., vol. 693, pp. 1–56, 2017
[3] E. G. Flekkøy, R. Delgado-Buscalioni, and P. V. Coveney, “Flux boundary conditions in particle simulations,” Phys. Rev. E, vol. 72, p. 026703, 2005
[4] https://zenodo.org/records/17257343
[5] P. Español, “Hydrodynamics from dissipative particle dynamics,” Phys. Rev. E, vol. 52, p. 1734, 1995